Section 2.6 — Inventory of continuous distributions¶
This notebook contains all the code examples from Section 2.6 Inventory of continuous distributions of the No Bullshit Guide to Statistics.
Notebook setup¶
# load Python modules
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
# Figures setup
sns.set_theme(
context="paper",
style="whitegrid",
palette="colorblind",
rc={'figure.figsize': (7,4)},
)
%config InlineBackend.figure_format = 'retina'
# set random seed for repeatability
np.random.seed(42)
%pip install ministats
Requirement already satisfied: ministats in /Users/ivan/Projects/Minireference/software/ministats (0.2.0) Requirement already satisfied: matplotlib>=3.8.3 in /Users/ivan/Projects/Minireference/STATSbook/noBSstatsnotebooks/venv/lib/python3.9/site-packages (from ministats) (3.8.3) Requirement already satisfied: numpy>=1.26.4 in /Users/ivan/Projects/Minireference/STATSbook/noBSstatsnotebooks/venv/lib/python3.9/site-packages (from ministats) (1.26.4) Requirement already satisfied: scipy>=1.12.0 in /Users/ivan/Projects/Minireference/STATSbook/noBSstatsnotebooks/venv/lib/python3.9/site-packages (from ministats) (1.12.0) Requirement already satisfied: pandas>=2.2.1 in /Users/ivan/Projects/Minireference/STATSbook/noBSstatsnotebooks/venv/lib/python3.9/site-packages (from ministats) (2.2.1) Requirement already satisfied: pillow>=10.2.0 in /Users/ivan/Projects/Minireference/STATSbook/noBSstatsnotebooks/venv/lib/python3.9/site-packages (from ministats) (10.2.0) Requirement already satisfied: seaborn>=0.13.2 in /Users/ivan/Projects/Minireference/STATSbook/noBSstatsnotebooks/venv/lib/python3.9/site-packages (from ministats) (0.13.2) Requirement already satisfied: statsmodels>=0.14.1 in /Users/ivan/Projects/Minireference/STATSbook/noBSstatsnotebooks/venv/lib/python3.9/site-packages (from ministats) (0.14.1) Requirement already satisfied: python-dateutil>=2.7 in /Users/ivan/Projects/Minireference/STATSbook/noBSstatsnotebooks/venv/lib/python3.9/site-packages (from matplotlib>=3.8.3->ministats) (2.8.2) Requirement already satisfied: importlib-resources>=3.2.0 in /Users/ivan/Projects/Minireference/STATSbook/noBSstatsnotebooks/venv/lib/python3.9/site-packages (from matplotlib>=3.8.3->ministats) (5.12.0) Requirement already satisfied: cycler>=0.10 in /Users/ivan/Projects/Minireference/STATSbook/noBSstatsnotebooks/venv/lib/python3.9/site-packages (from matplotlib>=3.8.3->ministats) (0.11.0) Requirement already satisfied: kiwisolver>=1.3.1 in /Users/ivan/Projects/Minireference/STATSbook/noBSstatsnotebooks/venv/lib/python3.9/site-packages (from matplotlib>=3.8.3->ministats) (1.4.4) Requirement already satisfied: fonttools>=4.22.0 in /Users/ivan/Projects/Minireference/STATSbook/noBSstatsnotebooks/venv/lib/python3.9/site-packages (from matplotlib>=3.8.3->ministats) (4.38.0) Requirement already satisfied: packaging>=20.0 in /Users/ivan/Projects/Minireference/STATSbook/noBSstatsnotebooks/venv/lib/python3.9/site-packages (from matplotlib>=3.8.3->ministats) (23.0) Requirement already satisfied: contourpy>=1.0.1 in /Users/ivan/Projects/Minireference/STATSbook/noBSstatsnotebooks/venv/lib/python3.9/site-packages (from matplotlib>=3.8.3->ministats) (1.0.7) Requirement already satisfied: pyparsing>=2.3.1 in /Users/ivan/Projects/Minireference/STATSbook/noBSstatsnotebooks/venv/lib/python3.9/site-packages (from matplotlib>=3.8.3->ministats) (3.0.9) Requirement already satisfied: tzdata>=2022.7 in /Users/ivan/Projects/Minireference/STATSbook/noBSstatsnotebooks/venv/lib/python3.9/site-packages (from pandas>=2.2.1->ministats) (2024.1) Requirement already satisfied: pytz>=2020.1 in /Users/ivan/Projects/Minireference/STATSbook/noBSstatsnotebooks/venv/lib/python3.9/site-packages (from pandas>=2.2.1->ministats) (2022.7.1) Requirement already satisfied: patsy>=0.5.4 in /Users/ivan/Projects/Minireference/STATSbook/noBSstatsnotebooks/venv/lib/python3.9/site-packages (from statsmodels>=0.14.1->ministats) (0.5.6) Requirement already satisfied: zipp>=3.1.0 in /Users/ivan/Projects/Minireference/STATSbook/noBSstatsnotebooks/venv/lib/python3.9/site-packages (from importlib-resources>=3.2.0->matplotlib>=3.8.3->ministats) (3.13.0) Requirement already satisfied: six in /Users/ivan/Projects/Minireference/STATSbook/noBSstatsnotebooks/venv/lib/python3.9/site-packages (from patsy>=0.5.4->statsmodels>=0.14.1->ministats) (1.16.0) [notice] A new release of pip is available: 23.0 -> 24.0 [notice] To update, run: pip3.9 install --upgrade pip Note: you may need to restart the kernel to use updated packages.
from ministats import plot_pdf
from ministats import plot_cdf
from ministats import plot_pdf_and_cdf
Review of formulas¶
Gamma function¶
from scipy.special import gamma as gammaf
gammaf(1) # = 0! = 1
1.0
gammaf(2) # = 1! = 1
1.0
gammaf(3) # = 2! = 2*1
2.0
gammaf(4) # = 3! = 3*2*1
6.0
gammaf(5) # = 4! = 4*3*2*1
24.0
[gammaf(z) for z in [4, 4.1, 4.5, 4.9, 5]]
[6.0, 6.812622863016677, 11.63172839656745, 20.66738596185786, 24.0]
# plot gammaf between 0 and 5
xs = np.linspace(0.05, 5, 1000)
fXs = gammaf(xs)
ax = sns.lineplot(x=xs, y=fXs, label="$\\Gamma(z)$")
ax.set_xlabel("z")
Text(0.5, 0, 'z')
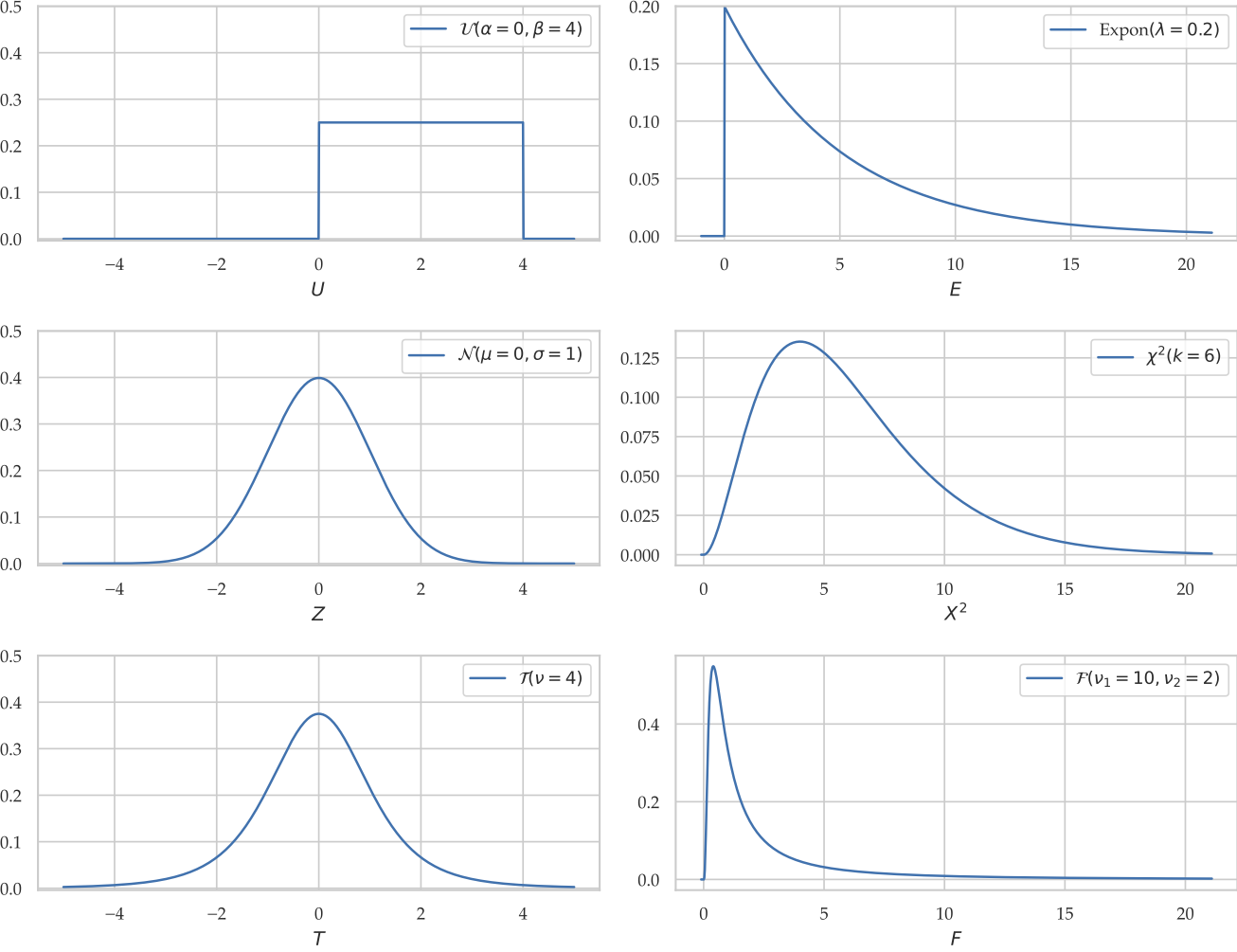
Continuous distribution reference¶
Uniform distribution¶
The uniform distribution $\mathcal{U}(\alpha,\beta)$ is described by the following probability density function:
$$ p_X(x) = \begin{cases} \frac{1}{\beta-\alpha} & \textrm{for } \alpha \leq x \leq \beta, \\ 0 & \textrm{for } x<0 \textrm{ or } x>1. \end{cases} $$
For a uniform distribution $\mathcal{U}(\alpha,\beta)$, each $x$ between $\alpha$ and $\beta$ is equally likely to occur, and values of $x$ outside this range have zero probability of occurring.
from scipy.stats import uniform
alpha = 2
beta = 7
rvU = uniform(alpha, beta-alpha)
# draw 10 random samples from X
rvU.rvs(10)
array([3.87270059, 6.75357153, 5.65996971, 4.99329242, 2.7800932 ,
2.7799726 , 2.29041806, 6.33088073, 5.00557506, 5.54036289])
plot_pdf(rvU, xlims=[0,9]);
# # ALT. use sns.lineplot
# # plot the probability density function (pdf) of the random variable X
# xs = np.linspace(0, 10, 1000)
# fUs = rvU.pdf(xs)
# sns.lineplot(x=xs, y=fUs)
Cumulative distribution function¶
plot_pdf_and_cdf(rvU, xlims=[0,9]);
Standard uniform distribution¶
The standard uniform distribution $U_s \sim \mathcal{U}(0,1)$ is described by the following probability density function:
$$ p_U(x) = \begin{cases} 1 & \textrm{for } 0 \leq x \leq 1, \\ 0 & \textrm{for } x<0 \textrm{ or } x>1. \end{cases} $$
where $U$ is the name of the random variable and $u$ are particular values it can take on.
The above equation describes tells you how likely it is to observe $\{U_s=x\}$. For a uniform distribution $\mathcal{U}(0,1)$, each $x$ between 0 and 1 is equally likely to occur, and values of $x$ outside this range have zero probability of occurring.
from scipy.stats import uniform
rvUs = uniform(0, 1)
# draw 10 random samples from X
rvUs.rvs(1)
array([0.02058449])
import random
random.seed(3)
random.random()
0.23796462709189137
random.uniform(0,1)
0.5442292252959519
import numpy as np
np.random.seed(42)
np.random.rand(10)
array([0.37454012, 0.95071431, 0.73199394, 0.59865848, 0.15601864,
0.15599452, 0.05808361, 0.86617615, 0.60111501, 0.70807258])
plot_pdf_and_cdf(rvUs, xlims=[-1,2]);
Simulating other random variables¶
We can use the uniform random variable to generate random variables from other distributions.
For example,
suppose we want to generate observations of a coin toss random variable
which comes out heads 50% of the time and tails 50% of the time.
We can use the standard uniform random variables obtained from random.random()
and split the outcomes at the "halfway point" of the sample space,
to generate the 50-50 randomness of a coin toss.
The function flip_coin defined below shows how to do this:
def flip_coin():
u = random.random() # random number in [0,1]
if u < 0.5:
return "heads"
else:
return "tails"
# simulate one coin toss
flip_coin()
'heads'
# simulate 10 coin tosses
[flip_coin() for i in range(0,10)]
['tails', 'tails', 'heads', 'heads', 'tails', 'heads', 'heads', 'tails', 'heads', 'tails']
Exponential¶
from scipy.stats import expon
lam = 7
loc = 0
scale = 1/lam
rvE = expon(loc, scale)
The computer model expon accepts as its first argument an optional "location" parameter,
which can shift the exponential distribution to the right,
but we want loc=0 to get the simple case,
that corresponds to the un-shifted distribution $\textrm{Expon}(\lambda)$.
rvE.mean(), rvE.var()
(0.14285714285714285, 0.02040816326530612)
# math formulas for mean and var
1/lam, 1/lam**2
(0.14285714285714285, 0.02040816326530612)
## ALT. we can obtain mean and ver using the .stats() method
## The code below also computes the skewness and the kurtosis
# mean, var, skew, kurt = rvE.stats(moments='mvsk')
# mean, var, skew, kurt
# f_E(5) = pdf value at x=10
rvE.pdf(0.2)
1.7261787475912451
plot_pdf(rvE, xlims=[0,1.1]);
Normal¶
A random variable $N$ with a normal distribution $\mathcal{N}(\mu,\sigma)$ is described by the probability density function:
$$ f_N(x) = \tfrac{1}{\sigma\sqrt{2\pi}} e^{\small -\tfrac{(x-\mu)^2}{2\sigma^2}}. $$
The mean $\mu$ and the standard deviation $\sigma$ are called the parameters of the distribution. The math notation $\mathcal{N}(\mu, \sigma)$ is used to describe the whole family of normal probability distributions.
from scipy.stats import norm
mu = 10 # = 𝜇 where is the centre?
sigma = 3 # = 𝜎 how spread out is it?
rvN = norm(mu, sigma)
rvN.mean(), rvN.var()
(10.0, 9.0)
plot_pdf(rvN, xlims=[-10,30]);
# ALT. generate the plot manually
# create a normal random variable
from scipy.stats import norm
mean = 1000 # 𝜇 (mu) = where is its center?
std = 100 # 𝜎 (sigma) = how spread out is it?
rvN = norm(mean, std)
# plot its probability density function (pdf)
xs = np.linspace(300, 1700, 1000)
ys = rvN.pdf(xs)
ax = sns.lineplot(x=xs, y=ys)
Standard normal¶
A standard normal is denoted $Z$ with a normal distribution $\mathcal{N}(\mu=0,\sigma=1)$ and described by the probability density function:
$$ f_Z(z) = \tfrac{1}{\sqrt{2\pi}} e^{\small -\tfrac{z^2}{2}}. $$
from scipy.stats import norm
rvZ = norm(0,1)
rvZ.mean(), rvZ.var()
(0.0, 1.0)
fig, ax = plt.subplots()
plot_pdf(rvZ, xlims=[-4,4], ax=ax, rv_name="Z")
<Axes: xlabel='z', ylabel='$f_{Z}$'>
Cumulative probabilities in the tails¶
Probability of $Z$ being smaller than $-2.2$.
rvZ.cdf(-2.3)
0.010724110021675809
Probability of $Z$ being greater than $2.2$.
1 - rvZ.cdf(2.3)
0.010724110021675837
Probability of $|Z| > 2.2$.
rvZ.cdf(-2.3) + (1-rvZ.cdf(2.3))
0.021448220043351646
norm.cdf(-2.3,0,1) + (1-norm.cdf(2.3,0,1))
0.021448220043351646
Inverse cumulative distribution calculations¶
rvZ.ppf(0.05)
-1.6448536269514729
rvZ.ppf(0.95)
1.6448536269514722
rvZ.interval(0.9)
(-1.6448536269514729, 1.6448536269514722)
Student's $t$-distribution¶
This is a generalization of the standard normal with "heavy" tails.
from scipy.stats import t
df = 10
rvT = t(df)
ax = plot_pdf(rvT, xlims=[-5,5], label=f"t({df})")
plot_pdf(rvZ, xlims=[-5,5], ax=ax, label="Z");
rvT.mean(), rvT.var()
(0.0, 1.25)
# Kurtosis formula kurt(rvT) = 6/(df-4) for df>4
rvT.stats("k")
1.0
rvT.cdf(-2.3)
0.022127156642143552
rvT.ppf(0.05), rvT.ppf(0.95)
(-1.8124611228107341, 1.8124611228107335)
fig, ax = plt.subplots()
linestyles = ['solid', 'dashdot', 'dashed', 'dotted']
for i, df in enumerate([2,5,10,100]):
rvT = t(df)
linestyle = linestyles[i]
plot_pdf(rvT, xlims=[-5,5], ax=ax, label="$\\nu={}$".format(df), linestyle=linestyle)
Snedecor's $F$-distribution¶
from scipy.stats import f
df1, df2 = 15, 10
rvF = f(df1, df2)
rvF.mean(), rvF.var()
(1.25, 0.7986111111111112)
plot_pdf(rvF, xlims=[0,5]);
Chi-squared distribution¶
from scipy.stats import chi2
k = 10
rvX2 = chi2(k)
rvX2.mean(), rvX2.var()
(10.0, 20.0)
1 - rvX2.cdf(20)
0.02925268807696113
plot_pdf(rvX2, xlims=[0,40]);
from scipy.stats import gamma as gammad
alpha = 4
loc = 0
lam = 2
beta = 1/lam
rvG = gammad(alpha, loc, beta)
rvG.mean(), rvG.var()
(2.0, 1.0)
plot_pdf(rvG, xlims=[0,5]);
Beta (optional)¶
from scipy.stats import beta as betad
alpha = 3
beta = 7
rvB = betad(alpha, beta)
rvB.mean(), rvB.var()
(0.3, 0.019090909090909092)
plot_pdf(rvB, xlims=[0,1]);
Cauchy (optional)¶
from scipy.stats import cauchy
x0 = 3
gamma = 5
rvC = cauchy(x0, gamma)
rvC.mean(), rvC.var()
(nan, nan)
plot_pdf(rvC, xlims=[-40,40]);