Section 4.3 — Interpreting linear models¶
This notebook contains the code examples from Section 4.3 Interpreting linear models from the No Bullshit Guide to Statistics.
Notebook setup¶
# load Python modules
import os
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
# Figures setup
plt.clf() # needed otherwise `sns.set_theme` doesn't work
from plot_helpers import RCPARAMS
# RCPARAMS.update({'figure.figsize': (8, 5)}) # good for screen
RCPARAMS.update({'figure.figsize': (5, 3)}) # good for print
# RCPARAMS.update({'text.latex.preamble': r'\usepackage{amsmath}'})
# RCPARAMS.update({'text.usetex': True})
sns.set_theme(
context="paper",
style="whitegrid",
palette="colorblind",
rc=RCPARAMS,
)
# High-resolution please
%config InlineBackend.figure_format = 'retina'
# Where to store figures
DESTDIR = "figures/lm/interpreting"
<Figure size 640x480 with 0 Axes>
# set random seed for repeatability
np.random.seed(42)
import statsmodels.formula.api as smf
import statsmodels.api as sm
import statsmodels.stats.api as sms
Introduction¶
# load the dataset
doctors = pd.read_csv("../datasets/doctors.csv")
n = doctors.shape[0]
# fit the model
formula = "score ~ 1 + alc + weed + exrc"
lm2 = smf.ols(formula, data=doctors).fit()
# model degrees of freedom (number of predictors)
p = lm2.df_model
# display the summary table
lm2.summary()
| Dep. Variable: | score | R-squared: | 0.842 |
|---|---|---|---|
| Model: | OLS | Adj. R-squared: | 0.839 |
| Method: | Least Squares | F-statistic: | 270.3 |
| Date: | Fri, 19 Jul 2024 | Prob (F-statistic): | 1.05e-60 |
| Time: | 10:45:38 | Log-Likelihood: | -547.63 |
| No. Observations: | 156 | AIC: | 1103. |
| Df Residuals: | 152 | BIC: | 1115. |
| Df Model: | 3 | ||
| Covariance Type: | nonrobust |
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | 60.4529 | 1.289 | 46.885 | 0.000 | 57.905 | 63.000 |
| alc | -1.8001 | 0.070 | -25.726 | 0.000 | -1.938 | -1.662 |
| weed | -1.0216 | 0.476 | -2.145 | 0.034 | -1.962 | -0.081 |
| exrc | 1.7683 | 0.138 | 12.809 | 0.000 | 1.496 | 2.041 |
| Omnibus: | 1.140 | Durbin-Watson: | 1.828 |
|---|---|---|---|
| Prob(Omnibus): | 0.565 | Jarque-Bera (JB): | 0.900 |
| Skew: | 0.182 | Prob(JB): | 0.638 |
| Kurtosis: | 3.075 | Cond. No. | 31.2 |
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
Model fit metrics¶
Coefficient of determination¶
lm2.rsquared
0.8421649167873537
# ALT.
1 - lm2.ssr/lm2.centered_tss
0.8421649167873537
Adjusted coefficient of determination¶
lm2.rsquared_adj
0.8390497506713147
# ALT.
1 - (lm2.ssr/(n-p-1)) / (lm2.centered_tss/(n-1))
0.8390497506713146
F-statistic and associated p-value¶
lm2.fvalue, lm2.f_pvalue
(270.34350189265825, 1.0512133413867047e-60)
Log-likelihood¶
lm2.llf
-547.6259042117637
# ALT.
from scipy.stats import norm
sigmahat = np.sqrt(lm2.scale)
np.sum(np.log(norm.pdf(lm2.resid,scale=sigmahat)))
# Q: Why not exactly the same as lm2.llf?
-547.651992151218
# How lm2.llf is computed:
SSR = np.sum(lm2.resid**2)
nobs2 = n/2.0
llf = -np.log(SSR)*nobs2 # concentrated likelihood
llf -= ( 1+np.log(np.pi/nobs2) )*nobs2 # with likelihood constant
llf
-547.6259042117637
Information criteria¶
lm2.aic, lm2.bic
(1103.2518084235273, 1115.4512324525256)
# ALT.
2*(p+1) - 2*lm2.llf, np.log(n)*(p+1) - 2*lm2.llf
(1103.2518084235273, 1115.4512324525256)
Other model-fit metrics¶
The mean squared error (MSE)
from statsmodels.tools.eval_measures import mse
scores = doctors["score"]
scorehats = lm2.fittedvalues
mse(scores,scorehats), lm2.ssr/n
(65.56013803717558, 65.56013803717559)
Parameter estimates¶
# estimated parameters
lm2.params
Intercept 60.452901 alc -1.800101 weed -1.021552 exrc 1.768289 dtype: float64
# estimated sigma (std. of error term)
sigmahat = np.sqrt(lm2.scale)
sigmahat
8.202768119825624
# ALT.
np.sqrt(lm2.ssr/(n-p-1))
8.202768119825624
Confidence intervals for model parameters¶
# standard errors
lm2.bse
Intercept 1.289380 alc 0.069973 weed 0.476166 exrc 0.138056 dtype: float64
lm2.conf_int(alpha=0.05)
| 0 | 1 | |
|---|---|---|
| Intercept | 57.905480 | 63.000321 |
| alc | -1.938347 | -1.661856 |
| weed | -1.962309 | -0.080794 |
| exrc | 1.495533 | 2.041044 |
Hypothesis testing for linear models¶
T-tests for individual parameters¶
Hypothesis testing for slope coefficient
Is there a non-zero slope coefficient?
Example: t-test for the variable weed¶
Null hypothesis $H_0$:
weedhas no effect onscore, which is equivalent to $\beta_{\texttt{weed}} = 0$:$$ H_0: \quad S \;\sim\; \mathcal{N}( {\color{red} \beta_0 + \beta_{\texttt{alc}}\!\cdot\!\textrm{alc} + \beta_{\texttt{exrc}}\!\cdot\!\textrm{exrc} }, \ \sigma) \qquad \qquad $$
Alternative hypothesis $H_A$:
weedhas an effect onscore, and the slope is not zero, $\beta_{\texttt{weed}} \neq 0$:$$ H_A: \quad S \;\sim\; \mathcal{N}\left( {\color{blue} \beta_0 + \beta_{\texttt{alc}}\!\cdot\!\textrm{alc} + \beta_{\texttt{weed}}\!\cdot\!\textrm{weed} + \beta_{\texttt{exrc}}\!\cdot\!\textrm{exrc} }, \ \sigma \right) $$
lm2.tvalues["weed"], lm2.pvalues["weed"], n-p-1
(-2.145370545436854, 0.033511561813423775, 152.0)
Calculate the $t$ statistic:
obst_weed = (lm2.params["weed"] - 0) / lm2.bse["weed"]
obst_weed # = lm2.tvalues["weed"]
-2.145370545436854
Calculate the associated $p$-value
from scipy.stats import t as tdist
pleft = tdist(df=n-p-1).cdf(obst_weed)
pright = 1 - tdist(df=lm2.df_resid).cdf(obst_weed)
pvalue_weed = 2 * min(pleft, pright)
pvalue_weed # = lm2.pvalues["weed"]
0.033511561813423775
lm2.tvalues
Intercept 46.885245 alc -25.725654 weed -2.145371 exrc 12.808529 dtype: float64
lm2.df_resid, n-p-1
(152.0, 152.0)
lm2.pvalues
Intercept 2.756807e-92 alc 2.985013e-57 weed 3.351156e-02 exrc 6.136296e-26 dtype: float64
F-test for the overall model¶
lm2.fvalue
270.34350189265825
# ALT.
F = lm2.mse_model / lm2.mse_resid
F
270.34350189265825
lm2.f_pvalue
1.0512133413867047e-60
# ALT.
from scipy.stats import f as fdist
fdist(dfn=p, dfd=n-p-1).sf(F)
1.0512133413867047e-60
F-test for a submodel¶
formula2nw = "score ~ 1 + alc + exrc"
lm2_noweed = smf.ols(formula2nw, data=doctors).fit()
F_noweed, p_noweed, _ = lm2.compare_f_test(lm2_noweed)
F_noweed, p_noweed
(4.602614777227957, 0.033511561813425135)
The F-statistic is the same as the t-statistic¶
F_noweed, obst_weed**2, lm2.tvalues["weed"]**2
(4.602614777227957, 4.602614777228025, 4.602614777228025)
p_noweed, pvalue_weed, lm2.pvalues["weed"]
(0.033511561813425135, 0.033511561813423775, 0.033511561813423775)
Assumptions checks and diagnostics¶
Are the assumptions for the linear model satisfied?
Residuals plots¶
from ministats import plot_resid
plot_resid(lm2, lowess=True);
from ministats import plot_resid
plot_resid(lm2, pred="alc", lowess=True);
Linearity checks¶
Look at residuals plots.
Independence checks¶
Look at residuals plots.
Normality checks¶
Look at residuals plots...
QQ plot inspection¶
from statsmodels.graphics.api import qqplot
qqplot(lm2.resid, line="s");
# BONUS
ax = sns.histplot(lm2.resid)
ax.set_xlim([-27,27]);
Skew and kurtosis¶
from scipy.stats import skew
from scipy.stats import kurtosis
skew(lm2.resid), kurtosis(lm2.resid, fisher=False)
(0.18224568453683496, 3.0747952385562636)
Homoscedasticity checks¶
The scale-location plot allows us to see if pattern in the variance of the residuals exists.
from ministats import plot_scaleloc
plot_scaleloc(lm2);
# ALT. Generate the scale-location plot from scratch
# 1. Compute the square root of the standardized residuals
scorehats = lm2.fittedvalues
std_resids = lm2.resid / np.sqrt(lm2.scale)
sqrt_std_resids = np.sqrt(np.abs(std_resids))
# 2. Generate the scatter plot
ax = sns.scatterplot(x=scorehats, y=sqrt_std_resids)
# 3. Add the LOWESS curve
from statsmodels.nonparametric.smoothers_lowess import lowess
xgrid, ylowess = lowess(sqrt_std_resids, scorehats).T
sns.lineplot(x=xgrid, y=ylowess, ax=ax)
# 4. Add descriptive labels
ax.set_ylabel(r"$\sqrt{|standardized\ residuals|}$")
ax.set_xlabel("fitted values");
Collinearity checks¶
Check the correlation matrix¶
corrM = doctors[["alc", "weed", "exrc"]].corr()
corrM.round(4)
| alc | weed | exrc | |
|---|---|---|---|
| alc | 1.0000 | 0.0422 | 0.0336 |
| weed | 0.0422 | 1.0000 | 0.0952 |
| exrc | 0.0336 | 0.0952 | 1.0000 |
# # BONUS 1 (not covered anywhere earlier in the book)
# sns.heatmap(corrM, annot=True, fmt='.2f', cmap="Grays");
# # BONUS 2 (not covered anywhere earlier in the book)
# sns.pairplot(doctors[["alc", "weed", "exrc"]]);
Condition number¶
lm2.condition_number
31.229721453770164
# ALT. (compute from scratch)
#######################################################
X = sm.add_constant(doctors[["alc","weed","exrc"]])
eigvals = np.linalg.eigvals(X.T @ X)
lam_max, lam_min = np.max(eigvals), np.min(eigvals)
np.sqrt(lam_max/lam_min)
31.229721453770264
Variance inflation factor¶
from ministats import calc_lm_vif
calc_lm_vif(lm2, "alc"), calc_lm_vif(lm2, "weed"), calc_lm_vif(lm2, "exrc")
(1.0026692156304757, 1.010703145007322, 1.010048809433661)
# ALT. using variance_inflation_factor from statsmodels
from statsmodels.stats.outliers_influence \
import variance_inflation_factor as statsmodels_vif
X = sm.add_constant(doctors[["alc","weed","exrc"]])
statsmodels_vif(X, 1) # variation inflation factor of "alc" in lm2
1.0026692156304757
Perfect multicollinearity example¶
doctorscol = doctors.copy()
doctorscol["alc2"] = doctors["alc"]
formula2col = "score ~ 1 + alc + alc2 + weed + exrc"
lm2col = smf.ols(formula2col, data=doctorscol).fit()
lm2col.params
Intercept 60.452901 alc -0.900051 alc2 -0.900051 weed -1.021552 exrc 1.768289 dtype: float64
calc_lm_vif(lm2col, "alc")
/Users/ivan/Projects/Minireference/software/ministats/ministats/linear_models.py:17: RuntimeWarning: divide by zero encountered in scalar divide vif = 1. / (1. - r_squared_i)
inf
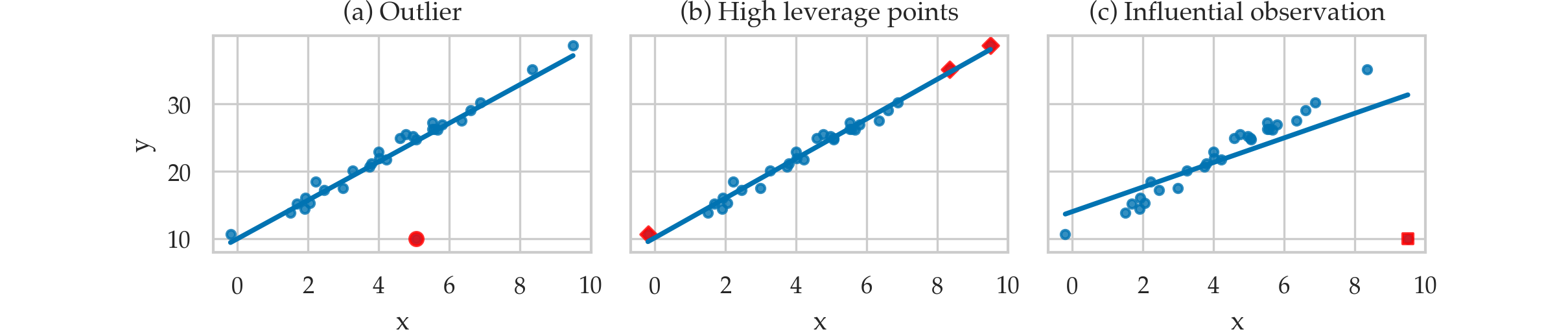
Leverage and influence metrics¶
TODO: add definitions of quantities in point form
Leverage plots¶
from statsmodels.graphics.api import plot_leverage_resid2
plot_leverage_resid2(lm2);
Influence plots¶
from statsmodels.graphics.api import influence_plot
influence_plot(lm2, criterion="cooks", size=4);
Model predictions¶
Prediction on the dataset¶
lm2.fittedvalues
0 55.345142
1 32.408174
2 71.030821
3 56.789073
4 57.514154
...
151 58.867105
152 43.049719
153 57.728460
154 69.617361
155 78.135788
Length: 156, dtype: float64
Prediction for new data¶
newdoc = {"alc":3, "weed":1, "exrc":8}
lm2.predict(newdoc)
0 68.177355 dtype: float64
newdoc_score_pred = lm2.get_prediction(newdoc)
newdoc_score_pred.conf_int(obs=True, alpha=0.1)
array([[54.50324518, 81.85146489]])
In-sample prediction accuracy¶
# Compute the in-sample mean squared error (MSE)
scores = doctors['score']
scorehats = lm2.fittedvalues
mse = np.mean( (scores - scorehats)**2 )
mse
65.56013803717558
# ALT.
lm2.ssr/n
65.56013803717559
# # ALT2.
# from statsmodels.tools.eval_measures import mse
# mse(scores,scorehats)
Out-of-sample prediction accuracy¶
TODO: explain
Leave-one-out cross-validation¶
loo_preds = np.zeros(n)
for i in range(n):
doctors_no_i = doctors.drop(index=i)
lm2_no_i = smf.ols(formula,data=doctors_no_i).fit()
predictors_i = dict(doctors.loc[i,:])
pred_i = lm2_no_i.predict(predictors_i)[0]
loo_preds[i] = pred_i
# Calculate the out-of-sample mean squared error
mse_loo = np.mean( (doctors['score'] - loo_preds)**2 )
mse_loo
69.20054514710948
Compare with the in-sample MSE of the model which is lower.
lm2.ssr/n
65.56013803717559
Towards machine learning¶
The out-of-sample prediction accuracy is a common metric used in machine learning (ML) tasks.
Explanations¶
Adjusted $R^2$¶
TODO: show formula
# TODO
Calculating standard error of parameters (optional)¶
lm2.bse
Intercept 1.289380 alc 0.069973 weed 0.476166 exrc 0.138056 dtype: float64
# construct the design matrix for the model
X = sm.add_constant(doctors[["alc","weed","exrc"]])
# calculate the diagonal of the inverse-covariance matrix
inv_covs = np.diag(np.linalg.inv(X.T.dot(X)))
sigmahat*np.sqrt(inv_covs)
array([1.28938008, 0.06997301, 0.4761656 , 0.13805557])
# these lead to approx. same result because predictors are not correlated
# but the correct formula is to use the inv. covariance matrix as above.
sum_alc_dev2 = np.sum((doctors["alc"] - doctors["alc"].mean())**2)
sum_weed_dev2 = np.sum((doctors["weed"] - doctors["weed"].mean())**2)
sum_exrc_dev2 = np.sum((doctors["exrc"] - doctors["exrc"].mean())**2)
se_b_alc = sigmahat / np.sqrt(sum_alc_dev2)
se_b_weed = sigmahat / np.sqrt(sum_weed_dev2)
se_b_exrc = sigmahat / np.sqrt(sum_exrc_dev2)
se_b_alc, se_b_weed, se_b_exrc
(0.06987980522527788, 0.47363764322385443, 0.13736710473639846)
Metrics for influential observations¶
Leverage score¶
$$ h_i = 1 - \frac{r_i}{r_{i(i)}}, $$
infl = lm2.get_influence()
infl.hat_matrix_diag[0:5]
array([0.09502413, 0.0129673 , 0.01709783, 0.01465484, 0.00981632])
Studentized residuals¶
$$ t_{i} = \frac{r_i}{\hat{\sigma}_{(i)} \sqrt{1-h_i}} $$
infl.resid_studentized_external[0:5]
array([ 0.98085317, -2.03409243, -1.61072775, -0.21903275, -1.29094028])
Cook's distance¶
$$ D_i = \frac{r_i^2}{p \, \widehat{\sigma}^2} \cdot \frac{h_{i}}{(1-h_{i})^2} % = \frac{t_i^2 }{p}\frac{h_i}{1-h_i} $$
infl.cooks_distance[0][0:5]
array([0.02526116, 0.01331454, 0.01116562, 0.00017951, 0.0041123 ])
DFFITS diagnostic¶
$$ \textrm{DFFITS}_i = \frac{ \widehat{y}_i - \widehat{y}_{i(i)} }{ \widehat{\sigma}_{(i)} \sqrt{h_i} }. $$
infl.dffits[0][0:5]
array([ 0.31783554, -0.23314692, -0.21244055, -0.02671194, -0.12853536])
Calculating leverage and influence metrics using statsmodels¶
infl = lm2.get_influence()
infl_df = infl.summary_frame()
# list(infl_df.columns)
cols = ["student_resid", "hat_diag", "cooks_d", "dffits"]
infl_df[cols].round(3).head(5)
| student_resid | hat_diag | cooks_d | dffits | |
|---|---|---|---|---|
| 0 | 0.981 | 0.095 | 0.025 | 0.318 |
| 1 | -2.034 | 0.013 | 0.013 | -0.233 |
| 2 | -1.611 | 0.017 | 0.011 | -0.212 |
| 3 | -0.219 | 0.015 | 0.000 | -0.027 |
| 4 | -1.291 | 0.010 | 0.004 | -0.129 |
Hypothesis tests for checking linear model assumptions¶
lm2.diagn
{'jb': 0.8999138579592632,
'jbpv': 0.6376556155083237,
'skew': 0.18224568453683496,
'kurtosis': 3.0747952385562636,
'omni': 1.1403999852814017,
'omnipv': 0.5654123490825906,
'condno': 31.229721453770164,
'mineigval': 40.14996367643264}
Hypothesis test for linearity¶
sms.linear_harvey_collier(lm2)
TtestResult(statistic=1.4317316989777105, pvalue=0.15427366179829397, df=152)
Normality tests¶
from statsmodels.stats.stattools import omni_normtest
omni, omnipv = omni_normtest(lm2.resid)
omni, omnipv
(1.1403999852814017, 0.5654123490825906)
from statsmodels.stats.stattools import jarque_bera
jb, jbpv, skew, kurtosis = jarque_bera(lm2.resid)
jb, jbpv, skew, kurtosis
(0.8999138579592632, 0.6376556155083237, 0.18224568453683496, 3.0747952385562636)
Independence test (autocorrelation)¶
from statsmodels.stats.stattools import durbin_watson
durbin_watson(lm2.resid)
1.8284830211766727
lm, p_lm, _, _ = sms.het_breuschpagan(lm2.resid, lm2.model.exog)
lm, p_lm
(6.254809752854315, 0.09985027888796541)
Goldfeld-Quandt test¶
# sort by score
idxs = doctors.sort_values("alc").index
# sort by scorehats
idxs = np.argsort(lm2.fittedvalues)
X = lm2.model.exog[idxs,:]
resid = lm2.resid[idxs]
# run the test
F, p_F, _ = sms.het_goldfeldquandt(resid, X, idx=1)
F, p_F
(1.5346515257083702, 0.03369473000286662)
Discussion¶
Maximum likelihood estimate¶
from scipy.optimize import minimize
from scipy.stats import norm
n = len(doctors)
preds = doctors[["alc", "weed", "exrc"]]
scores = doctors["score"]
def negloglikelihood(betas):
scorehats = betas[0] + (betas[1:]*preds).sum(1)
residuals = scores - scorehats
SSR = np.sum(residuals**2)
sigmahat = np.sqrt(SSR/(n-3-1))
likelihoods = norm.pdf(residuals,scale=sigmahat)
loglikelihood = np.sum(np.log(likelihoods))
return -1 * loglikelihood
betas = minimize(negloglikelihood, x0=[0,0,0,0]).x
betas
array([60.4528995 , -1.80010127, -1.02155223, 1.76828879])
lm2.params.values
array([60.45290059, -1.80010132, -1.02155166, 1.76828876])
Regularization¶
doctors[["alc", "weed", "exrc"]].std()
alc 9.428506 weed 1.391068 exrc 4.796361 dtype: float64
# TRUE
# alc = - 1.8
# weed = - 0.5
# exrc = + 1.9
# FITTED
lm2.params
Intercept 60.452901 alc -1.800101 weed -1.021552 exrc 1.768289 dtype: float64
L1 regularization (LASSO)¶
Set alpha option to the desired value $\alpha_1$ and L1_wt = 1,
which means $100\%$ L1 regularization.
lm2_L1reg = smf.ols(formula, data=doctors) \
.fit_regularized(alpha=0.3, L1_wt=1.0)
lm2_L1reg.params
Intercept 58.930367 alc -1.763306 weed 0.000000 exrc 1.795228 dtype: float64
L2 regularization (ridge)¶
Set alpha option to the desired value $\alpha_2$ and L1_wt = 0,
which means $0\%$ L1 regularization and $100\%$ L2 regularization.
lm2_L2reg = smf.ols(formula, data=doctors) \
.fit_regularized(alpha=0.05, L1_wt=0.0001)
lm2_L2reg.params
Intercept 50.694960 alc -1.472536 weed -0.457425 exrc 2.323386 dtype: float64
Statsmodels diagnostics plots (BONUS TOPIC)¶
TODO: import explanations from .tex
Plot fit against one regressor¶
see https://www.statsmodels.org/stable/generated/statsmodels.graphics.regressionplots.plot_fit.html
from statsmodels.graphics.api import plot_fit
with plt.rc_context({"figure.figsize":(9,2.6)}):
fig, (ax1,ax2,ax3) = plt.subplots(1,3, sharey=True)
plot_fit(lm2, "alc", ax=ax1)
plot_fit(lm2, "weed", ax=ax2)
plot_fit(lm2, "exrc", ax=ax3)
ax2.get_legend().remove()
ax3.get_legend().remove()
from statsmodels.graphics.api import plot_partregress
with plt.rc_context({"figure.figsize":(9,2.6)}):
fig, (ax1,ax2,ax3) = plt.subplots(1,3, sharey=True)
plot_partregress("score", "alc", exog_others=[], data=doctors, obs_labels=False, ax=ax1)
plot_partregress("score", "weed", exog_others=[], data=doctors, obs_labels=False, ax=ax2)
plot_partregress("score", "exrc", exog_others=[], data=doctors, obs_labels=False, ax=ax3)
CCPR plot¶
component and component-plus-residual
see https://www.statsmodels.org/stable/generated/statsmodels.graphics.regressionplots.plot_ccpr.html
from statsmodels.graphics.api import plot_ccpr
with plt.rc_context({"figure.figsize":(9,2.6), "text.usetex":True}):
fig, (ax1,ax2,ax3) = plt.subplots(1,3)
fig.subplots_adjust(wspace=0.5)
plot_ccpr(lm2, "alc", ax=ax1)
ax1.set_title("")
ax1.set_ylabel(r"$\texttt{alc} \cdot \widehat{\beta}_{\texttt{alc}} \; + \; \mathbf{r}$")
plot_ccpr(lm2, "weed", ax=ax2)
ax2.set_title("")
ax2.set_ylabel(r"$\texttt{weed} \cdot \widehat{\beta}_{\texttt{weed}} \; + \; \mathbf{r}$", labelpad=-3)
plot_ccpr(lm2, "exrc", ax=ax3)
ax3.set_title("")
ax3.set_ylabel(r"$\texttt{exrc} \cdot \widehat{\beta}_{\texttt{exrc}} \; + \; \mathbf{r}$", labelpad=-3)
All-in-on convenience method¶
from statsmodels.graphics.api import plot_regress_exog
with plt.rc_context({"figure.figsize":(9,6)}):
plot_regress_exog(lm2, "alc")
Exercises¶
E4.LOGNORM Non-normal error term¶
from scipy.stats import uniform, lognorm
np.random.seed(42)
xs = uniform(0,10).rvs(100)
ys = 2*xs + lognorm(1).rvs(100)
df = pd.DataFrame({"x":xs, "y":ys})
lm = smf.ols("y ~ x", data=df).fit()
qqplot(lm.resid, line="s");
# sns.scatterplot(x=xs, y=lm.resid);
# from ministats import plot_lm_scale_loc
# plot_lm_scale_loc(lm);
E4.DEP Dependent error term¶
E4.NL Nonlinear relationship¶
from scipy.stats import uniform, lognorm
np.random.seed(42)
xs = uniform(0,10).rvs(100)
ys = 2*xs + xs**2 + norm(0,1).rvs(100)
df = pd.DataFrame({"x":xs, "y":ys})
lm = smf.ols("y ~ 1 + x", data=df).fit()
lm.params
Intercept -14.684585 x 11.688188 dtype: float64
from ministats import plot_reg
plot_reg(lm);
plot_resid(lm, lowess=True);
qqplot(lm.resid, line="s");
E4.H Heteroskedasticity¶
from scipy.stats import uniform, norm
np.random.seed(43)
xs = np.sort(uniform(0,10).rvs(100))
sigmas = np.linspace(1,20,100)
ys = 2*xs + norm(loc=0,scale=sigmas).rvs(100)
df = pd.DataFrame({"x":xs, "y":ys})
lm = smf.ols("y ~ x", data=df).fit()
from ministats import plot_scaleloc
plot_scaleloc(lm);
E4.v Collinearity¶
from scipy.stats import uniform, norm
np.random.seed(45)
x1s = uniform(0,10).rvs(100)
alpha = 0.8
x2s = alpha*x1s + (1-alpha)*uniform(0,10).rvs(100)
ys = 2*x1s + 3*x2s + norm(0,1).rvs(100)
df = pd.DataFrame({"x1":x1s, "x2":x2s, "y":ys})
lm = smf.ols("y ~ x1 + x2", data=df).fit()
lm.params, lm.bse
(Intercept 0.220927 x1 1.985774 x2 3.006225 dtype: float64, Intercept 0.253988 x1 0.137253 x2 0.167492 dtype: float64)
lm.condition_number
24.291898486251146
calc_lm_vif(lm, "x1"), calc_lm_vif(lm, "x2")
(17.17244980334442, 17.17244980334442)
Links¶
- More details about model checking https://ethanweed.github.io/pythonbook/05.04-regression.html#model-checking
- Statistical Modeling: The Two Cultures paper that explains the importance of out-of-sample predictions for statistical modelling.
https://projecteuclid.org/journals/statistical-science/volume-16/issue-3/Statistical-Modeling--The-Two-Cultures-with-comments-and-a/10.1214/ss/1009213726.full
CUT MATERIAL¶
Example dataset¶
mtcars = sm.datasets.get_rdataset("mtcars", "datasets").data
mtcars
lmcars = smf.ols("mpg ~ hp", data=mtcars).fit()
sns.scatterplot(x=mtcars["hp"], y=mtcars["mpg"])
sns.lineplot(x=mtcars["hp"], y=lmcars.fittedvalues);
Compute the variance/covariance matrix¶
lm2.cov_params()
# == lm2.scale * np.linalg.inv(X.T @ X)
# where X = sm.add_constant(doctors[["alc","weed","exrc"]])
| Intercept | alc | weed | exrc | |
|---|---|---|---|---|
| Intercept | 1.662501 | -0.055601 | -0.093711 | -0.095403 |
| alc | -0.055601 | 0.004896 | -0.001305 | -0.000288 |
| weed | -0.093711 | -0.001305 | 0.226734 | -0.006177 |
| exrc | -0.095403 | -0.000288 | -0.006177 | 0.019059 |